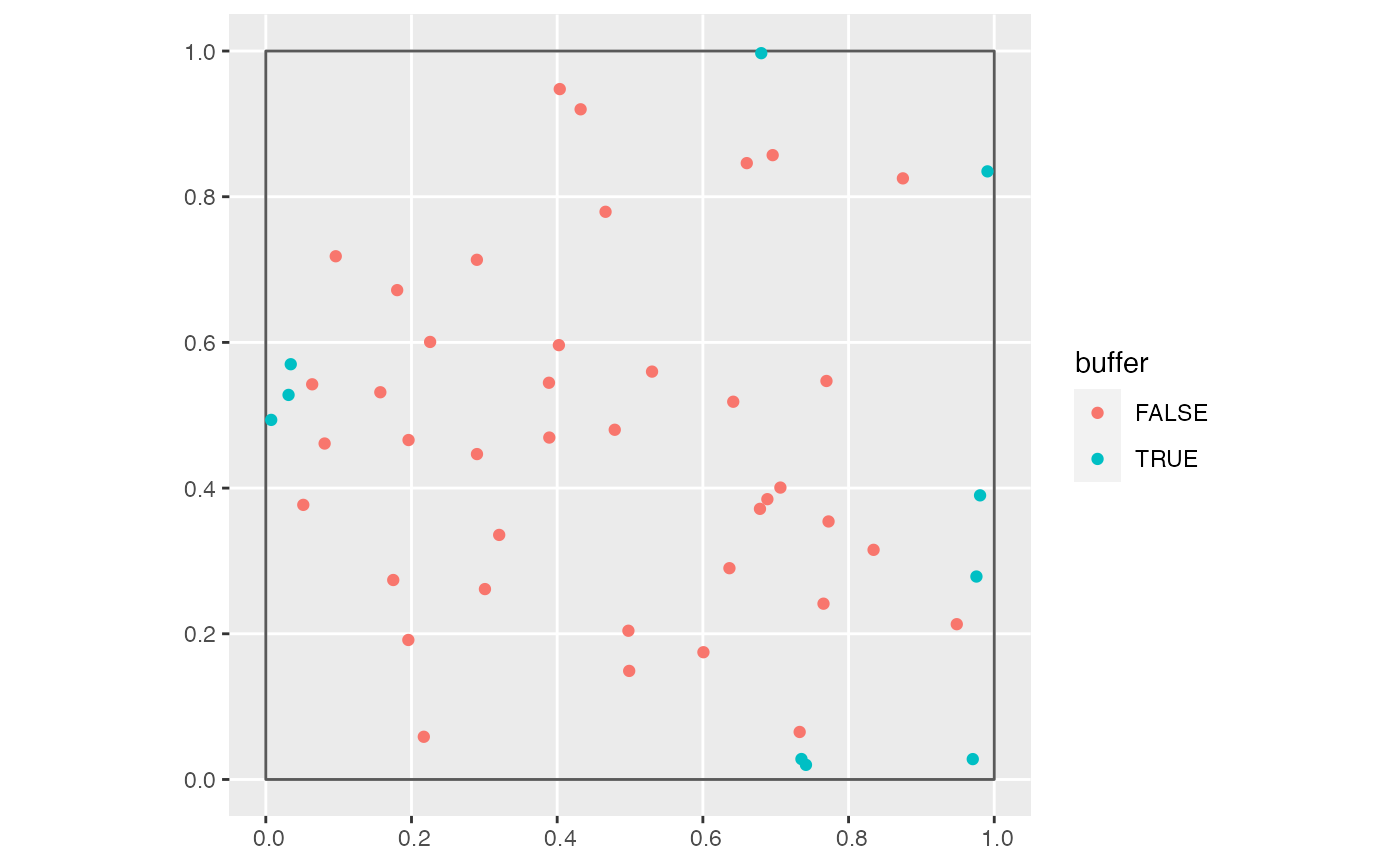
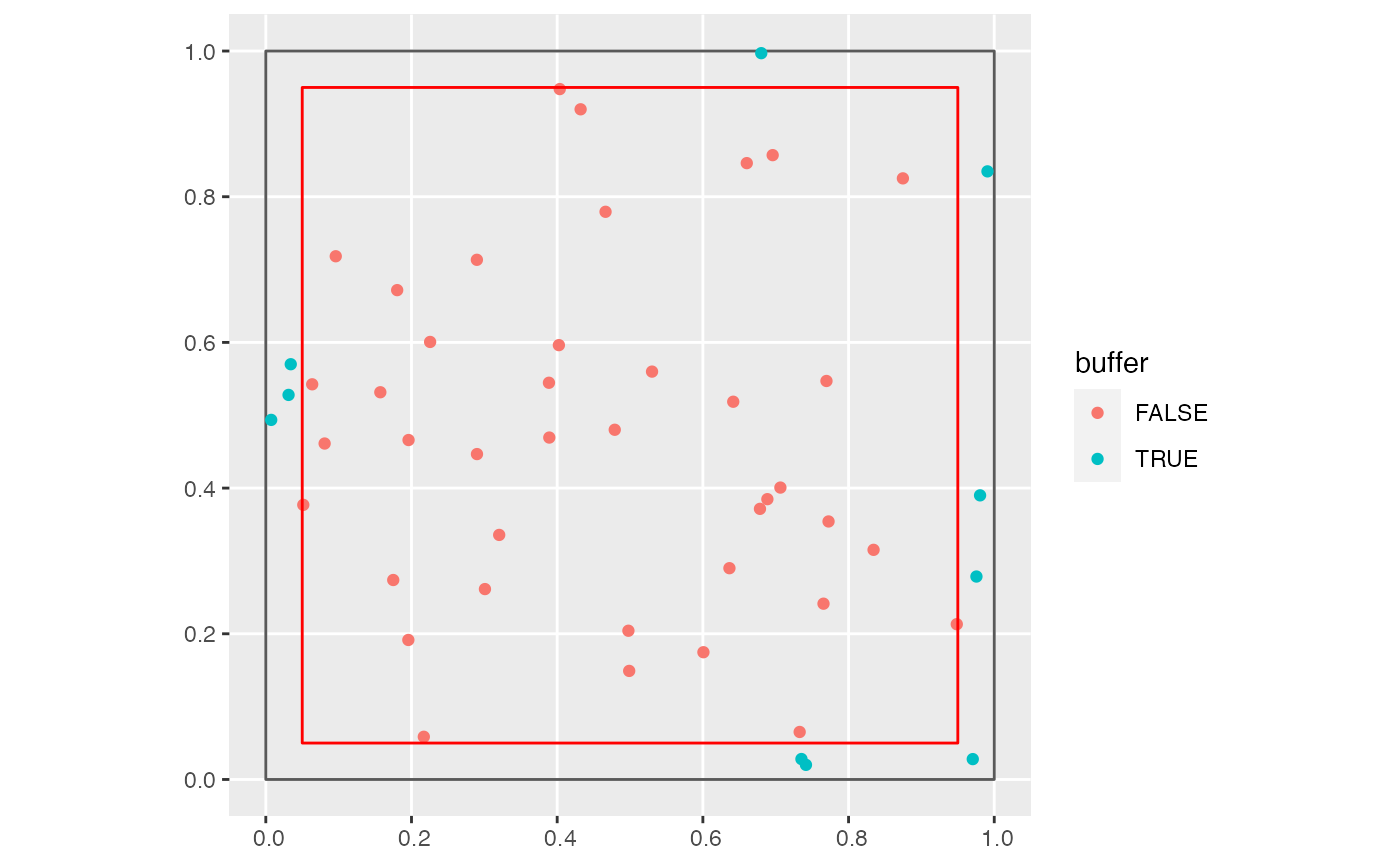
Identify trees in the buffer region
add_buffer_variable(growth_df, direction = "in", size, region)
Arguments
| growth_df |
|
|---|---|
| direction | "in" for buffers that are contained within |
| size | Distance to determine which neighboring trees to a focal tree are
competitors. The units are assumed to be the same as the |
| region | An |
Value
The same growth_df data frame but with a new boolean
variable buffer indicating if a tree is in the study region buffer
area. This uses compute_buffer_region() to define the boundary
of the buffer region.
See also
Other spatial functions:
compute_buffer_region(),
focal_vs_comp_distance()
Examples
library(tibble) library(sfheaders) library(ggplot2) # Example square region to be buffered region <- tibble( x = c(0, 0, 1, 1), y = c(0, 1, 1, 0) ) %>% sf_polygon() # Example points study_points <- tibble( x = runif(50), y = runif(50) ) %>% sf_point() # Size of buffer size <- 0.05 # Identify whether points are within size of boundary study_points <- study_points %>% add_buffer_variable(direction = "in", size = size, region = region) # Plot study points coded by whether they are within size of boundary p <- ggplot() + geom_sf(data = region, fill = "transparent") + geom_sf(data = study_points, aes(col = buffer)) p# Additionally, show buffer boundary in red buffer_boundary <- region %>% compute_buffer_region(direction = "in", size = size) p + geom_sf(data = buffer_boundary, col = "red", fill = "transparent")